Custom database tables for Ingest
You can ingest catalog data from your database by adding a custom database table for CATENTRY and CATGROUP. You can also control how database data is serialized into Elasticsearch to bring in large datasets.
Procedure
-
Set the following NiFi variables at the
ReindexLink,NRTLink, andDataloadLinkpipes.- Use custom.table.catentry to provide a custom CATENTRY table to refine the base scope of catalog entry SQLs.
- Use custom.where.catentry to provide a custom Where clause of the custom CATENTRY table.
- Use custom.table.catgroup to provide a custom CATGROUP table to refine the base scope of catalog group SQLs.
- Use custom.where.catgroup to provide a custom Where
clause of the above custom CATGROUP table.Note: once a custom table is defined, an equivalent set of TI_DELTA flowfile variables will be generated. The similar variable name all starts with X_CUSTOM instead of TI_DELTA.
Use these "custom table" and "custom condition" variables to generate an INNER JOIN condition for all of the SQLs in the default pipelines.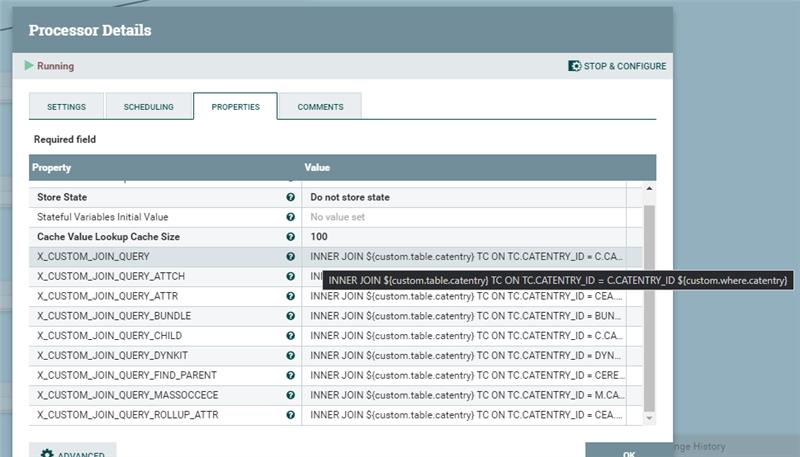 Use the X_CUSTOM_JOIN_QUERY variable in the SQL declared in each of the connector pipes. For example,
Use the X_CUSTOM_JOIN_QUERY variable in the SQL declared in each of the connector pipes. For example,
-
Optional: If you want to allow overriding of the database schema name, follow
the previous steps to update the NiFi variables in
ReindexLink,NRTLink, andDataloadLink. However, use flow.database.schema to define the custom database schema name to be used for indexing.